Google’s research team have open-sourced a new visualisation technology that allows researchers to view petabyte-scale 3D models of brains in a web browser.
The Neuroglancer project, available on Github, enables neurologists to build 3D models of a brain’s neural pathways in interactive visualisations.
Using a WebGL-based viewer for volumetric data, it can display non axis-aligned cross-sectional views of volumetric data, 3-D meshes and line-segment based models.
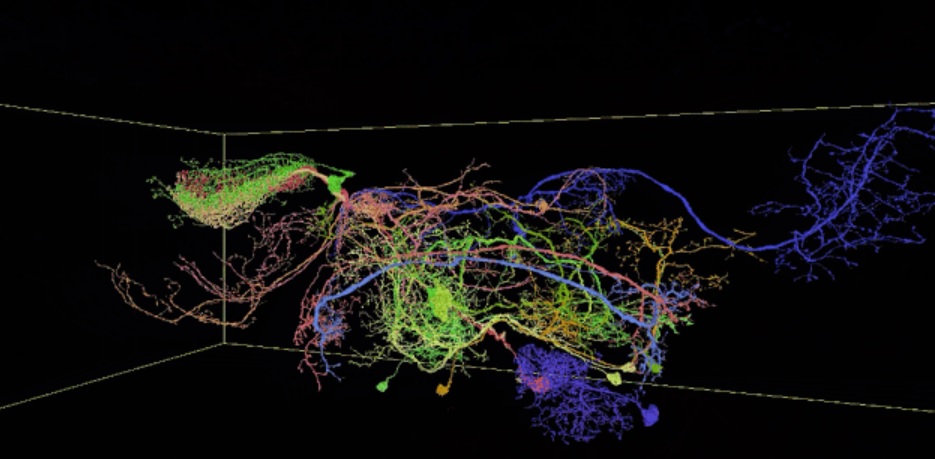
The Neuroglancer project (“not an official Google project”, its contributors note) has been used by scientists to create an interactive 3D interface of the brain of a fruit fly.
The humble fruit fly has long been instrumental in neural and genetic research (earning scientists eight Nobel prizes), thanks to a short life cycle which allows researchers to experiment on several generations of flies in a short span of time.
Importantly for neurological work, fruit flies have small brains containing only one hundred thousand neurons, in comparison to humans, which are equipped with over one hundred billion neurons.
Working with machine learning and neural networks; scientists were able to map the brain’s wiring in a fruit fly, which was then collated into an interactive map facilitated by the Neuroglancer project software.

Brain Mapping Visualization Technology
In order to map the fly’s neurons, researchers at Howard Hughes Medical Institute sliced a fruit fly’s brain into thousands of ultra-thin 40-nanometer sections that were then imaged using a transmission electron microscope, this process created a forty trillion pixel image of the brain.
Using Google’s Cloud TPU v3 Pods, (racks of Google’s Tensor Processing Units), the team used Flood-Filling Networks to automatically trace each individual neuron in the fly’s brain. Flood-Filling Networks are a new technique created by Google that combines the imaging capabilities of two algorithms.
Viren Jain, research scientist in Google’s Connectomics department, earlier described their use in a blog, saying: “Traditional algorithms have divided the process into at least two steps: finding boundaries between neurites using an edge detector or a machine-learning classifier, and then grouping together image pixels that are not separated by a boundary using an algorithm like watershed or graph cut.”
In 2015, they created a new approach that begins with a specific pixel location and then through an iteration process the algorithm, using a recurrent convolutional neural network, predicts which pixels are part of the same line or object.

The result is a trillion pixel interactive 3D image of the fruit fly’s brain which is viewable via the Neuroglancer software in any web browser that supports WebGL.
The team of researchers who wrote the scientific paper recording their work mapping the fly’s brain wrote that it: “Produced a largely merger-free segmentation of the entire ssTEM Drosophila brain (fruit fly), which we make freely available. As compared to manual tracing using an efficient skeletonization strategy, the segmentation enabled circuit reconstruction and analysis workflows that were an order of magnitude faster.”
The ability to browse full human brains in 3D remains some way off, but the tool may well prove useful for other enterprise data visualisation projects. The full interactive map is viewable here via the Neuroglancer software.






